Another excellent conference organised jointly by SCI and the RSC at the MSD Site in Oss, Netherlands. The third in the series featured a mixture of perspectives, case histories and new methods that gave an excellent view of the current state of GPCR drug discovery.
The conference started with Ad IJzermann presenting his group's A2A receptor crystal structure in complex with ZM241385 (PDB code 3EML). Ad went on to describe differences between published homology models and the experimentally determined structure (none of the homology models were close) and also virtual screening experiments that have been published using the A2A structure. Interestingly this topic was also covered on the last day by Malcolm Weir from Heptares where their alternative method for producing stabilised receptors gave a significantly different conformation for the ligand. It will be interesting to see the differences in more detail and compare the known ligands against each of these structures if and when the Heptares coordinates become available.
Anette Graven Sams from Lundbeck outlined their discovery of Lu AA47070, the prodrug from of Lu AA41063. There were several interesting points raised by Anette but the Chemist in me focussed on the introduction of a phosphate ester as a solubilising prodrug of an amino-thiazole. The group was introduced in high yield through sodium hydride deprotonation followed by quenching with chloromethyl chloroformate. I found it surprising that the deprotonation seemed to go exclusively and cleanly on the amino-thiazole nitrogen and was intrigued as to the mechanism of reaction, presumably some sort of imidate acts as the intermediate here (see scheme).
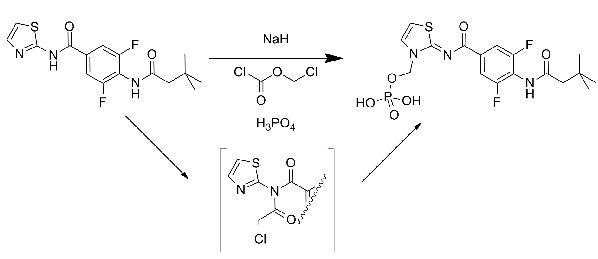
Anette showed the result of the docking of AA41063 into the 3EML A2A structure. She speculated on the requirement for a water molecule inside the binding pocket that led to the ligand adopting a position that was slightly 'higher' (closer to the extracellular edge) than for ZM241385. Pondering this, I wondered if the electronics of the central aromatic ring could be driving the binding of the Lundbeck compound as ZM241385 has an unusual face-face arrangement between the central triazotriazine and a phenylalanine (Phe168 in 3EML).
Excellent case histories from Brian Smith at Arena, Tseuneo Yasuma at Takeda and Sue Westerway at GSK highlighted the multi-optimisation problem of modern drug discovery, especially when working on a CNS located GPCR. The Takeda group had taken an interesting approach in modelling the natural ligand for GPR40 using a ligand based design strategy, something I am pretty sure the late Sir James Black would have approved of. The compounds that they produced were fairly long and stringy with log Ds in the 2-3 range. Still high for an acid but a lot lower than I would have expected given the natural substrate is a free fatty acid!
Mark Andrews from Pfizer showed an interesting piece of SAR. They had developed a range of basic pyrolidines as MC4 anatgonists but sought alternatives with better PK. They came up wth a set of pyrolidines substituted with heteroaromatics that showed a very specific SAR trend. All the aromatics required an asymmetric substitution pattern with respect to the position of the heteroatoms:
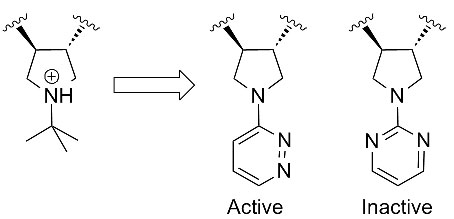
This kind of tight SAR seems quite unusual since the doubly substituted aromatics like the pyrimidine shown (above right) should benefit from reduced entropy over the single substitutions. Clearly there is a specific interaction here and it looks like aromatic C-Hs from the heteroaromatics mimic the protonated NH of the parent pyrolidine.
Steve Watson from GSK gave an interesting talk on the development of mGLuR5 negative allosteric modulators. Steve focussed his talk on looking at the brain free fraction when considering SAR. He argued that unless you consider the amount of the compound that can access the site of action at the same time as considering potency then you will take your research into blind alleys. Steve gave some excellent examples of how looking at the combination of potency and brain free fraction could drive you SAR. One extra thing that I noticed from the structures that he presented was that their final compound seemed to have a reasonable similarity to some of the previously reported examples when you consider Fields. I drew an Addex compound (presented by Steve) into FieldAlign to use as the reference structure, then draw GSK2210875 as a database molecule. The resulting alignment is shown below.
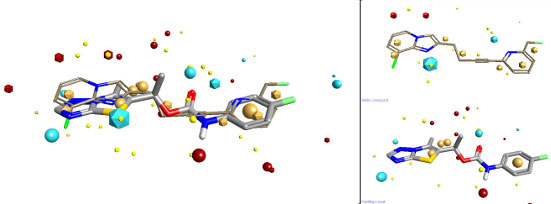
You can download a FieldView project file with all the structures mentioned in this review. FieldView is Cresset's free molecule viewer and editor.
I presented a poster on using FieldStere to find bioisosteres for MPEP, the prototype mGluyR5 allosteric modulator.
Tim Cheeseright, Cresset
