FEATURE BY ANTHONY KING | 8 NOVEMBER 2023
How does antibiotic resistance emerge? Anthony King discovers that the conventional narrative doesn't tell the whole story.
Antimicrobial resistance (AMR) is not a new phenomenon. Surprisingly, a 2016 study in the Lechuguilla Cave in New Mexico found a species of bacteria living 1,000ft underground that is resistant to most modern antibiotics[1] – including daptomycin, an antibiotic of so-called last resort often prescribed where other antibiotics no longer work. This is despite the fact the microbes in this limestone cave have been isolated for millions of years.

The microbes in Lechuguilla Cave, New Mexico, have been isolated for millions of years – yet it is home to a species of bacteria that is resistant to most modern antibiotics.
The same group, led by Gerard Wright at McMaster University in Canada, also discovered antibiotic resistance genes in permafrost in Yukon that was 30,000 years old.
‘Antibiotic resistance does not always come from the clinic,’ says William Gaze, Professor of microbiology at the University of Exeter, UK. ‘There are resistance genes in environmental bacteria that have existed for millions if not billions of years.’
Wild resistance
Escherichia coli undergoing the process of DNA exchange known as bacterial conjugation.Credit: Jonasz Patkowski
AMR was linked to an estimated 5m or so deaths in 2019, and this could reach 10m deaths/year by 2050, leading to calls for more R&D into new antibiotics. Beyond human and veterinary medicine, however, there is ‘growing evidence the environment plays a key role in the development, transmission and spread of AMR,’ as a 2023 UN Bracing for Superbugs report observes. Indeed, the EU is currently considering harmonising monitoring of AMR in the environment, including a network of national reference labs.
‘The consensus now is that most genes that confer resistance to medically important antibiotics have probably been acquired from existing microbiology,’ says Edward Topp, AMR scientist at the French National Institute for Agriculture, Food and Environment (INRAE). Considering that Alexander Fleming discovered penicillin in 1928, it makes sense that resistance to antimicrobial compounds pre-dates modern medicine.
David Graham at the University of Newcastle, UK, and one of the authors of the UN superbug report, points to two paths that resistance can follow. Bacteria stressed by living in a soup of lethal and non-lethal compounds, including antibiotics, can be pushed to mutate their existing machinery. A study of resistance to the antibiotic ciprofloxacin in S. aureus, for example, revealed that a few spontaneous mutations improved the actions of an efflux pump that flushed toxins out of the cell[2]. Physicians often focus on this pathway to resistance, since it can render antibiotics ineffective, and it is within their powers to restrict inappropriate use of antibiotics.

A 2023 study found bird excrement to be an especially rich source of resistance genes.
But the second path, says Graham, is the transmission and physical spread of resistance in the wild. A 2023 Swedish study, for example, revealed huge numbers of suspected latent resistance genes in different environments[3]. Wastewater and bird excrement were especially rich. ‘There is a large reservoir of resistance genes in the microbial world that is not associated with a pig or a human and has probably been there for billions of years,’ says Topp. ‘What we need to worry about is when bacteria reach out to get these genes in a way that subverts the efficacy of our medicines.’
Thankfully, the gap between the environment and people means it is not inevitable that resistance in the wild will get into clinically relevant microbes. But which route resistance takes is often obscure, as it remains challenging to trace the origins of AMR in a human patient. ‘It is very difficult to look at AMR in the clinic and say this came from food, or this percentage came from the environment,’ says Topp.
Pollution matters
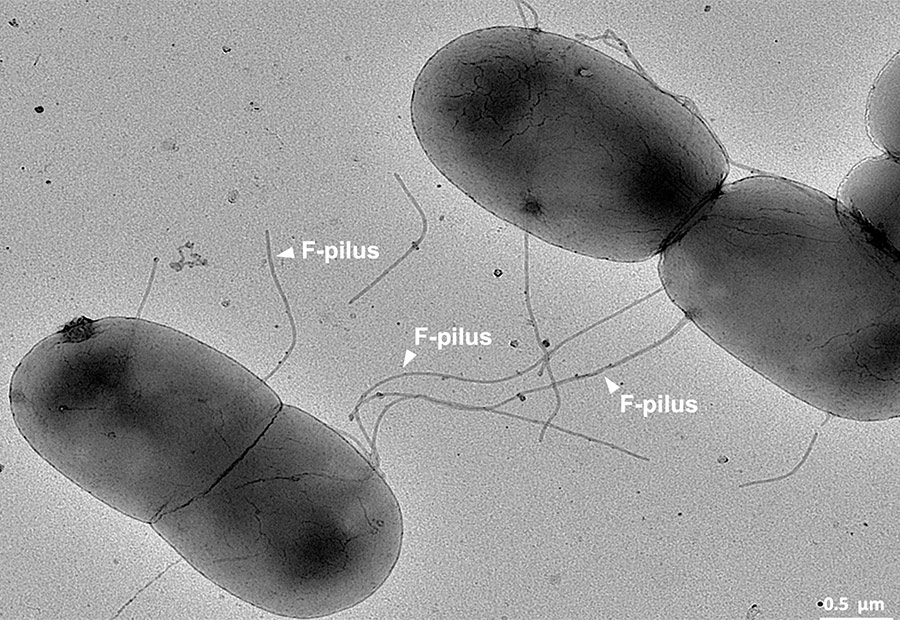
Escherichia coli undergoing the process of DNA exchange known as bacterial conjugation. Credit: Jonasz Patkowski
Increasing scrutiny of the environment as a source of resistance is occurring in tandem with rising support for the ‘One Health’ viewpoint, whereby animals, people and the environment are intrinsically connected, including the microbial world. AMR experts say that anthropogenic actions are stoking up background resistance in the environment and elevating risks that our gut bacteria will take it onboard. It is not just antibiotics in the environment that matters. Pollution, including everyday chemicals used at home, are also implicated.
To understand why, one needs to take a diversion into the weird world of microbial sex. ‘Bacteria can pass huge molecules of DNA between one another, even between completely unrelated species,’ says Gaze. Bacterial conjugation allows one strain to transfer DNA to another via a direct connection. This can involve swapping plasmids, which are circular pieces of DNA outside the chromosome. Such transfer has been implicated in the rise of resistance to a variety of beta-lactam antibiotics, including penicillins and carbapenems[4]. Viruses can also move resistance genes between bacteria, and sometimes microbes can excrete DNA into the environment, allowing other strains to gain resistance genes.
Certain bacteria seem more promiscuous in sharing DNA, while other microorganisms are permissive, readily accepting new DNA. But even if a soil- or water-borne bacterium contains resistance to multiple antibiotics, it still needs a reason to maintain this resistance, which usually comes at a metabolic cost, and a route to get into people. This is where two types of ubiquitous pollution enter the story.
First, although antibiotic inputs into waterways from manufacturing and poorly treated wastewater is unwelcome, it is misguided to think about only antibiotic pollution. The UN report lists fungicides, antiviral compounds, certain disinfectant chemicals and biocides such as zinc and copper as putting selective pressure on microbes to promote resistance spread. Reducing pollution is crucial to reducing the rise of superbugs, the UN said.
‘We use almost unimaginable amounts of biocides, antiseptics, detergents and fabric softeners that have antimicrobial properties,’ says Gaze, and this milieu pressures microbes in the environment into hitting their SOS response to toxic chemicals. This in turn means that mutation and gene transfer rates go up.
The second type of pollution we are releasing is human and animal waste. Releasing such bacteria plays a role in the spread, transfer and dissemination of resistance to antibiotics, say experts. Pollutants and gut bacteria are brought together in sewage treatment plants; treatment greatly reduces outflows of bacteria and AMR. Yet in parts of the world untreated sewage flows directly into waterways, including the UK during storm overflows. Longer contact times between chemicals and wastewater with indigenous microorganisms ‘increase the likelihood of AMR selection and transmission events’, the UN report notes.
As Graham points out: ‘There’s nothing that causes bacteria stress that can’t potentially cause bacteria to try and change genetically. The endpoint of almost any type of pollution is the development of slightly stronger microorganisms.’
Our gut bacteria are a special case. They are adapted for a topsy-turvy world, with irregular influxes of nutrients or antimicrobial compounds, and genetic flexibility is their superpower. ‘Organisms in the gut appear to be more genetically promiscuous than organisms in the environment,’ says Graham. He namechecks Escherichia coli, a rod-shaped bacterium that usually lives harmlessly in people, as being extraordinarily diverse. ‘They’re a little more hotwired to exchange and acquire genes,’ he says. While they can live as part of our normal flora, they can cause opportunistic infections; most E. coli strains go unnoticed in our gut, but rogue E. coli can cause urinary tract infections, meningitis and blood poisoning.
Many AMR researchers view the gut microbiome – whether harmless microbes, or potential pathogens – as critical in resistance spread. And while it remains difficult to track where a single resistance gene came from, there is a strong suspicion that food and water are the major routes into the human body.
Food and water

Johan Bengtsson-Palme. Credit: Chalmers
Precisely how microbes with resistance in their DNA get into people is a huge quandary. ‘It is important to figure out what are the dissemination routes to humans from the environment,’ says Johan Bengtsson-Palme, a microbial ecologist at Chalmers University of Technology in Gothenburg, Sweden. He advocates surveying the environment for resistance elements and then evaluating the risk that they can get into people. Once resistance gains access to human pathogens, ‘then it is just a selection and dissemination process, and there’s very little we can do about that’, says Bengtsson-Palme.
There are hints that exposure via food is especially important. The UK Food Standards Agency estimates there are around 2.4m cases of foodborne illness in the UK each year. ‘We don’t know about ingestion of antibiotic genes carried on benign bacteria,’ says Topp. ‘We have not even considered AMR in the food safety realm yet,’ he adds, with unsafe AMR ingestion levels unknown.
In 2018, a US study revealed that a strain of E. coli in poultry (ST131) is present in some human urinary tract infections. The lineage was present in poultry flocks since the 1940s. Further investigation suggested that this multi-drug resistant E. coli strain is infecting people from poultry meat[5].
Separately, resistance to colistin, one of the last remaining effective options against carbapenem-resistant Enterobacteriaceae, was first detected in 2015. Within a few years of the discovery of the resistance gene, mcr-1, it had spread across all continents. Sequencing data suggest that: ‘aquatic organisms are now recognised as a primary reservoir of mcr-1’, the UN report notes. Around 5% of healthy Dutch travellers to southern Africa and Asia acquired the mcr-1 gene; these people had no prior exposure to colistin.
But it is not just animals or meat products. Graham has been investigating AMR and infectious disease spread in informal settlements across New Delhi in India where water and food is often contaminated with human wastes. ‘Food is probably the biggest worry in these New Delhi communities, and one reason is that people don’t expect that,’ says Graham. ‘They don’t realise that the tomato or lettuce that they’ve got from the market has been exposed to nasty bacteria,’ possibly due to unsafe hygiene practices or exposure to water with faecal matter or sewage.

It is well known that the use of wastewater, sewage and manure on crops can lead to the spread of AMR.
A previous study of 45 neighbourhoods in ten cities around the world revealed that exposure to faecal matter via water was site specific and limited, whereas food was always the main route of exposure in low-income and middle-income cities[6]. It is well known that the use of wastewater, sewage and manure on crops can lead to the spread of AMR, while resistant microbes can also be enriched in soils irrigated with reused wastewater[7]. The UN report suggests that considerations of AMR should take account of crop types, such as whether it is likely to be eaten raw, when deciding to add such wastes to soil.
While medical doctors remain focused on the small number of bacteria that cause human infections, ‘they’re just the tip of the iceberg of antimicrobial resistance, which develops within microbial populations across humans, animals and the environmental microbiome’, says Gaze. He is convinced that food will be a major route of resistance transmission, arguing that antibiotic stewardship from the medical profession is not enough.
‘Globally, AMR has an enormous impact in terms of deaths, and it is escalating as a problem,’ says Maarten van Dongen, the Founder of AMR Insights, an organisation set up to combat and raise awareness of AMR. ‘In the past, AMR was really related to the human health sector, but then it was realised we see the same bacteria in chicken and pigs. Increasingly, we realise that the environment also plays a role.’
This is especially the case when you bring together human or animal gut bacteria in an environment where they are put under pressure from antimicrobials and in a position to tap into existing resistance reservoirs. This chimes with the UN report’s conclusion that better wastewater treatment, incentives to reduce antimicrobial use and regulation of discharges into the environment will play a part in suppressing the spread of superbugs. We know pollution is bad for AMR. We know sewage is bad. We know AMR will cause more needless deaths, says Gaze. ‘We should take a precautionary approach and reduce the spread of resistance.’
References
- Nature Commun., doi: 0.1038/ncomms13803
- Nature Commun., doi: 10.1038/s41467-020-17735-y
- Microbiome, doi: 10.1186/s40168-023-01479-0
- The ISME Journal, doi: 10.1038/s41396-023-01393-1
- mBio, doi: 10.1128/mbio.00470-18
- Science of the Total Environment, doi: 10.1016/j.scitotenv.2021.151273
- Water Research, doi: 10.1016/j.watres.2019.114906






